Pool-Seq
Time series study of Lumbricus terrestris
In TrendDNA, we make use of long-term sampling by the German Environmental Specimen Bank (ESB) across several locations in Germany over the last three decades. In this project, we analyze homogenate samples of the commonly known earthworm, Lumbricus terrestris, collected for most locations on a yearly basis from 1990 to 2021. This extensive sampling scheme provides a unique opportunity to detect significant selective changes in the earthworm genomes over this comprehensive time period and potentially identify causes of the observed genomic changes. To do so, it is first necessary to have a reference genome for the species.
Genome assembly
We first assemble the earthworm genome of an individual collected in Nidda Park (Frankfurt am Main) in spring 2022. Following the correct identification of the individual by taxonomic experts and barcoding approaches, we use Next-Generation Sequencing (NGS), a massively parallel sequencing approach that allows for assessing whole genomes with very high accuracy. Following sequencing, we use state-of-the-art bioinformatic software tools to obtain the complete earthworm genome up to the chromosome level. We use the assembled genome as a reference genome for the time series analyses.
Time series analyses
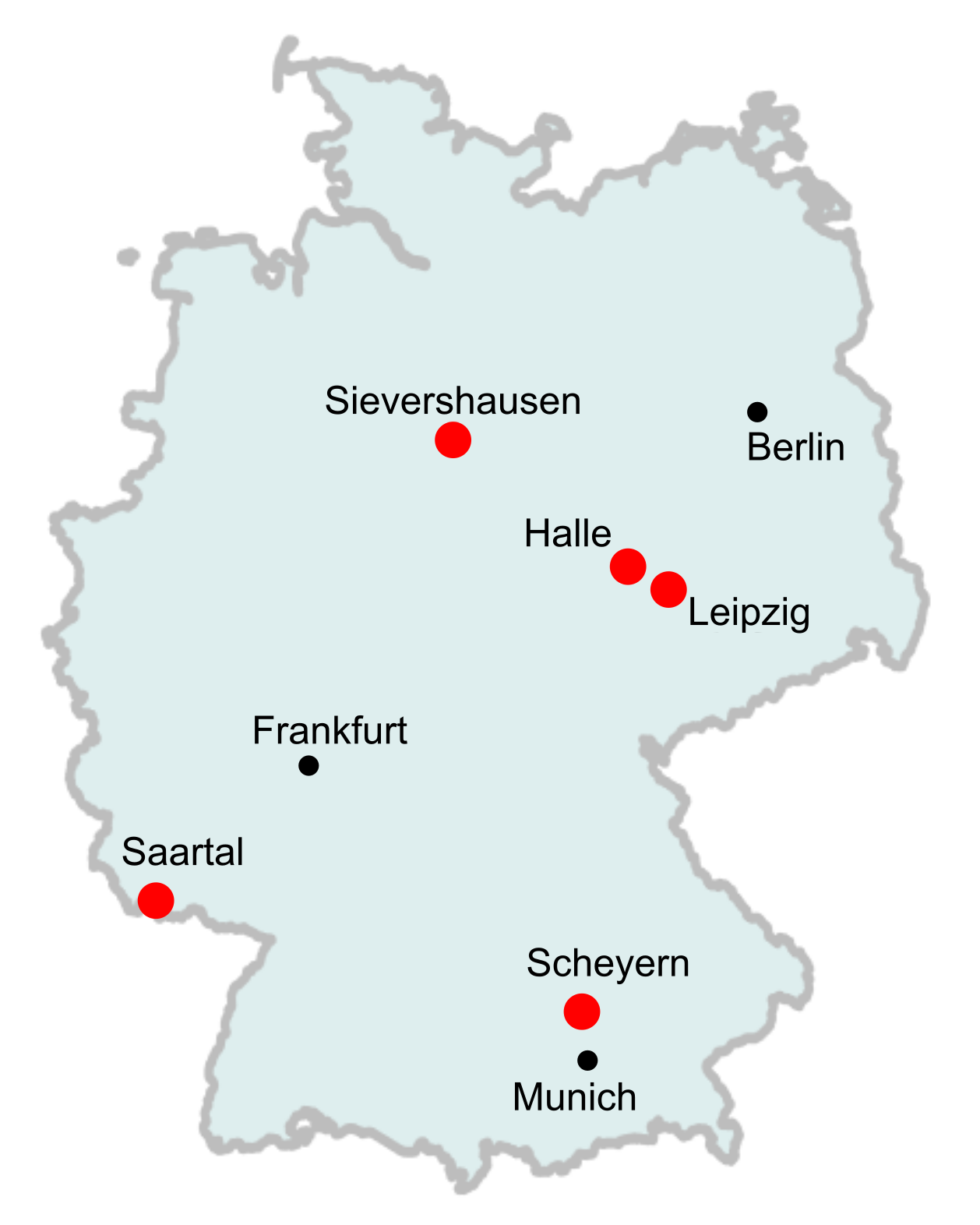
We use five ESB long-term sampling locations: Leipzig, Halle, Saartal, Scheyern and Sievershausen (Fig. 1). These locations cover a broad and variable geographical range, capturing a diversity of different environmental conditions across Germany. At each location, samples were collected at regular intervals during autumn and kept at -80 o C. Instead of sequencing each worm individually, which would be very costly and labor-intensive, we pool many individuals together, making sure that each one contributes approximately the same amount of tissue to the pool. Because of this equal contribution, we sequence each genetic variant according its true frequency in the population. By pooling samples, we thus obtain accurate estimates of numerous population genomic parameters. In total, this study includes 90 samples across the different locations, each sample containing up to 100 homogenized individuals. The amount of change over time will inform us whether evolutionary selection has occurred in the respective populations.