Conference and workshops on molecular methods for environmental observations, 26-28.03.2025, Essen
In 2025, the TrendDNA project will take part in a three-day DACHLiLux conference on the topic of ‘genetic methods in environmental monitoring’. The third day (28.03.2025) will focus on the works of the German environmental specimen bank during the TrendDNA project. We would like to redirect everyone interested towards the official webpage of the Umweltbundesamt, where you can register for the conference. The link below will lead you to the webpage.
We hope to welcome you, present and discuss our results.
https://www.umweltbundesamt.de/tagung-workshops-zu-molekularen-methoden-in-der
May 2024
The IUCN published a Statement regarding the use of eDNA methods for conservation and wildlife management. This signals the increasing usage and importance for environmental DNA as non-invasive and effective biomonitoring tool.

On May 25 th 2024, Florian Leese presented status quo and visions on biodiversity monitoring using DNA and eDNA based methods at an invited keynote talk at the 5 th National Symposium on eDNA bioassessments in Shenzhen, China.


At a „Sustainability“ science slam in Mainz, Florian Leese presented “Biodiversity – or the end of variety” on the 11 th of April 2024.


November 2023, Fraunhofer IME
WildOMICs is featured in the newsletter. The initiative strives to incorporate OMIC techniques in environmental monitoring, such as meta-transcriptomics.

September 2023
Isabelle Junk, Nina Schmitt and Henrik Krehenwinkel published a research article in the scientific journal Current Biology. The members of TrendDNA successfully tested blue mussel homogenate of the German ESB project for their ability to store environmental DNA in their associated communities through time and space. The authors were able to reconstruct the invasion trajectory of an barnacle.
18 June 2023
The Emschergenossenschaft and the NABU NRW invited researchers to the First Day of the living Emscher. The set goal was to record the diversity of species in the new mouth area of the once most polluted river in Germany. The TrendDNA-Team of the University of Duisburg-Essen took part in the event. Robin Schütz and Florian Leese, as well as Till-Hendrik Macher (GeDNA-Project) analyzed traces of animal species from environmental DNA in the water. Using a portable DNA-sequencing system, processing of the samples happened in the field. Identification through environmental DNA helped to discover numerous fish species, among others the eel and the ninespine stickleback.

24-25 April 2023
TrendDNA participants Jan Koschorreck (UBA), Henrik Krehenwinkel, Julian Hans and Isa Junk (Universität Trier), Robin Schütz (Universität Duisburg-Essen), and Bernd Göckener (Fraunhofer IME) give a progress report and project outlook at the UPB Jahrestagung in Berlin.

13-15 March 2023
Exciting new data from TrendDNA are presented at the 3rd Global Soil Biodiversity Conference in Dublin, Ireland. Here, Masters student Judith Paetsch (Senckenberg Gesellschaft, LOEWE-Zentrum-TBG) shows the lastest results of her project
using shotgun metagenomics to characterize the biodiversity of soil communities across space and time.
December 2022
Our study on arthropod biomonitoring using eDNA from tree leaves is now published open-access in eLife! In this innovative study led by Prof. Henrik Krehenwinkel of Universität Trier, UPB samples were used to measure trends in diversity and relative biomass of canopy arthropods over a 30-year period. While species richness remained relatively stable at individual sites, the species composition changed drastically, resulting in an overall homogenization of forest arthropod communities across Germany.
2-3 November 2022
TrendDNA members gather at the Fraunhofer IME for a thought-provoking discussion of possible future research directions and a tour of the Specimen Bank:



27-28 September 2022, University of Duisburg-Essen
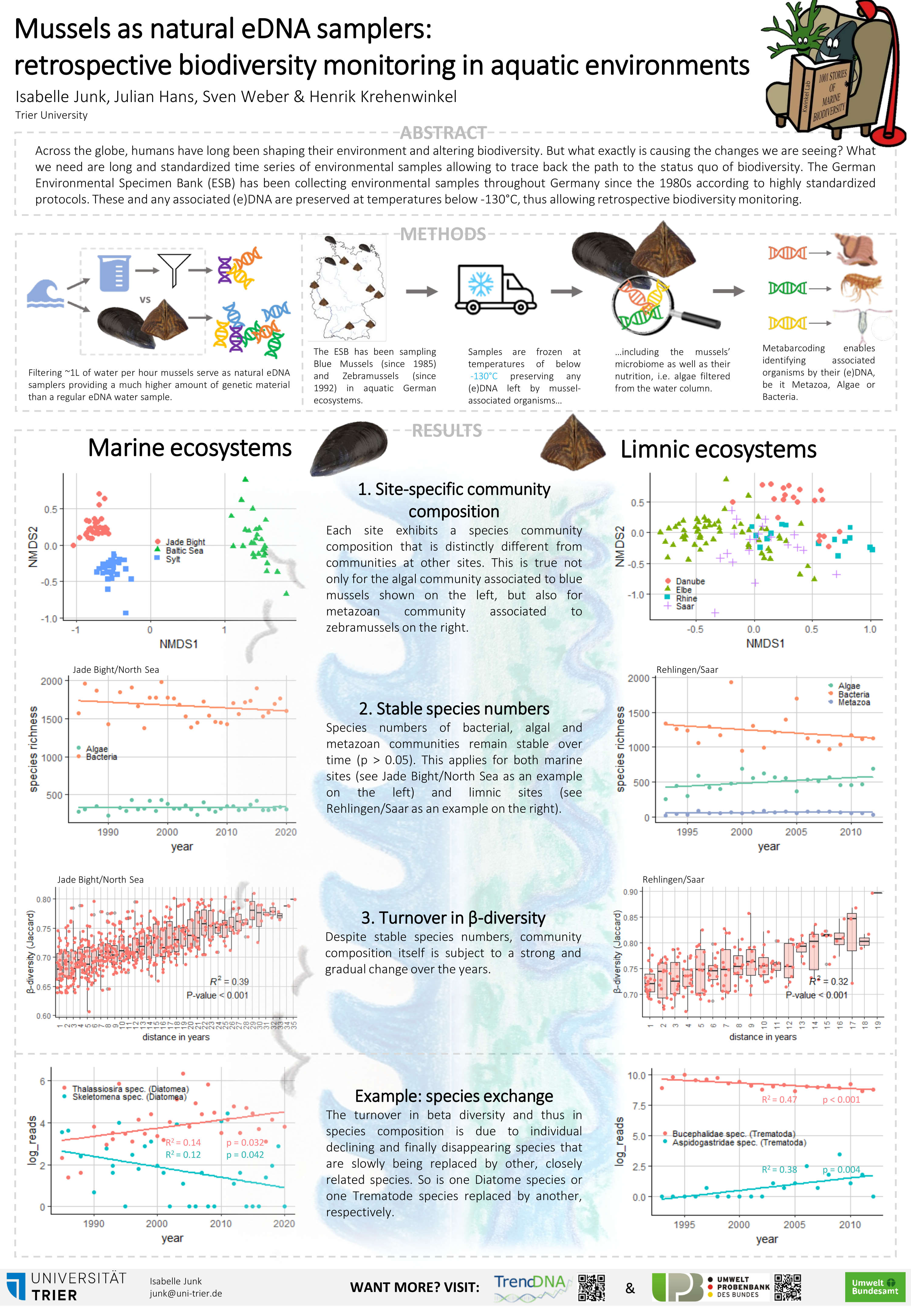
PhD students Robin Schütz (Universität Duisburg-Essen) and Isabelle Junk (Universität Trier) present posters of their research showing the exciting potential for eDNA metabarcoding of suspended particulate matter and mussels to reveal changes in limnic and marine communities over time:
Summer 2022
A companion project to TrendDNA is funded by the Bauer-Stiftung zur Förderung von Wissenschaft und Forschung. Led by Dr. Susan Kennedy, Prof. Henrik Krehenwinkel and Prof. Christoph Emmerling (Universität Trier), and in collaboration with Prof. Florian Leese (Universität Duisburg Essen), Prof. Miklós Bálint (Senckenberg TBG Frankfurt) and Prof. Thomas Udelhoven (Universität Trier), this exciting project uses metabarcoding, metagenomics and metatranscriptomics of earthworms to reveal changes in soil communities over time and across geographic distances.
See our Twitter page for more updates!